Resources
 Part of the Oxford Instruments Group
Part of the Oxford Instruments Group
Expand
Collapse
 Part of the Oxford Instruments Group
Part of the Oxford Instruments Group
Recently, with pandemics such as SARS, MERS and Covid-19, biosensors for the detection of viral pathogens have attracted a lot of interest. The rapid identification of the presence of a virus in contaminated food or water, surfaces or in a patients samples is a prerequisite to efficiently counteract viral outbreaks, epidemics as well as bioterrorism.
Currently, the highest sensitivity for virus detection is achieved with assays relying on polymerase chain reaction (PCR) for multiplication and detection of viral DNA and RNA. However, this technique has some drawbacks. It has a long processing time (typically 24 hours), the lack of real-time monitoring and rapid on-site pathogen detection and as well as this advanced laboratory equipment and trained personnel is required.
A research group led by Fredrik Höök at Chalmers University of Technology in Sweden have developed a bio-analytical assay for the detection of whole virus particles with single virus sensitivity. In their recent work, their focus is on the detection of the human Norovirus (NoV); a highly infectious virus causing gastroenteritis or the more commonly known “winter vomiting bug”.
Virus-like particles (VLPs) of the most common Norovirus II.4 genogroup (Ast/6139/01 strain) were used as models for the pathogen in this study. Norovirus is a small non-enveloped RNA virus of the Caliciviridae family with a high genetic diversity. It consists of an outer icosahedral shell assembled mainly from 180 capsid proteins which protect the viral genome.
The “winter vomiting bug” caused by this virus is spread globally through pandemics. Although it does not get the attention of other pandemics such as MERS or SARS, is responsible for over 200,000 annual deaths mainly in children in developing countries. The very stable and extremely infectious Norovirus spreads through the oral-faecal route and spontaneous outbreaks often occur after consumption of contaminated food or water. Its surveillance is of most importance since very low particle numbers (<100) may be sufficient to generate disease outbreaks, therefore making its early detection challenging.
Self-assembled capsid proteins recombinantly expressed in insect cells are often used to probe the binding behaviour of the virus. These non-infectious virus-like proteins (VLPs) exhibit a morphology and binding properties similar to those of the real virus and recognise with high specificity a variety of saliva and cell surface glycoconjugates, including membrane bound histo-blood group active glycosyphingolipids (GSL) . Such VLPs represent excellent models in work aimed at designing new biosensor principles for virus detection.
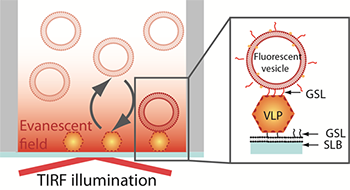
Figure 1. Schematic representation of the sandwich assay setup. The particles are captured onto a bilayer containing 10 % H type I glycosphingolipid (GSL) recognising the VLPs with high specificity. The fluorescence signal is generated TIRF illumination of the sensor-bound rhodamine-labeled vesicles containing 5% H type 1GSL.
The assay explored in this study is based on a sandwich-type configuration where the VLPs are first captured onto a non-fouling (low non-specific adsorption of proteins) supported lipid bilayer containing a NoV specific GSL-ligand. The firmly bound VLPs are then visualized by imaging individual fluorescently-labelled phospholipid vesicles containing the same NoV-specific ligand (Figure 1). Taking advantage of the evanescent field generated by TIRF illumination to discriminate surface-bound vesicles from those in solution, vesicle binding and release events at the sensor interface can be determined and tracked. This setup makes it therefore not only possible to visualize individual virus particles but also to gain information on their affinity to a receptor, the GSLs in this case. [1]
Time-lapse movies consisting of 1000 frames were acquired using TIRF microscopy combined with an iXon EMCCD camera at a rate of 5 frames per second. The assay was done in a 96-well plate and all wells of interest were imaged in 7 different positions to ensure reproducible statistics. A MatLab-based software was used to analyse the images. Briefly, a fluorescent vesicle was counted if its intensity exceeded a pre-set threshold and if it was present on a pre-defined number of consecutive frames (minimum 7 frames). Association plots were generated by recording the number of newly arrived vesicles over time and the residence time of the vesicles was also measured. The limit of detection of the assay was also quantified.
The results from this study show that the biosensor developed by Dr. Höök and his colleagues for the detection of the norovirus exhibited a good specificity with little non-specific binding (Figure 2). The method exhibits single molecule sensitivity as individual vesicles can be readily visualized. The vesicles generate localized signals which can be easily resolved and discriminated from the background noise. While background noise can be efficiently suppressed, a critical factor influencing the limit of detection (LOD) is the background signal generated by non-specific binding events. An important aspect to be taken into consideration is the non-fouling character of the sensing interface which should be virus and vesicle repellent. The lipid bilayers used in this study fulfilled these conditions. Additionally, the authors show how one can take advantage of the real-time monitoring of vesicle binding events to discriminated specific form non-specific binding, according to the vesicle residence time on the substrate and therefore to further decrease the non-specific background signal.
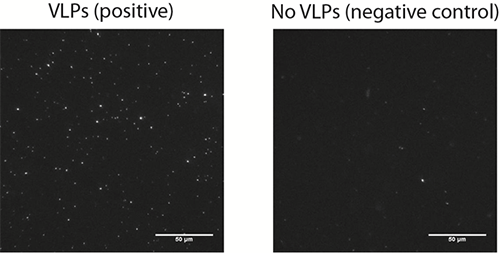
Figure 2. A representative microscopy image of surface bound vesicles (left image) on a bilayer incubated with 12.5 pM VLPs and on a negative control (right image) performed in the absence of VLPs.
The research group have demonstrated that a relatively simple assay setup combined with single-molecule sensitivity and equilibrium fluctuation analysis can be used to detect viral particles with a LOD of ~106 particles/ml. This LOD is better than what has so far been reported by others in the context of biosensors for the detection of noroviruses and additionally the whole assay could be performed in less than two hours a significant improvement on the current most sensitive method of virus detection where the PCR is employed. Furthermore, these techniques could be extended to study of other viral pathogens.
[1] Bally, M., Gunnarsson, A.; Svensson, L.; Larson, G. Zhdanov; V. P.; Höök, F., Interaction of Single Virus-like Particles with Vesicles Containing Glycosphingolipids. Physical Review Letters, 107 (18) pp. 188103.
The iXon EMCCD Life is the ideal camera for TIRF microscopy. The latest iXon 888 model is the fastest EMCCD detector available, reading out at 93 fps at 512 x 512 and 26 fps for the full 1024 x 1024 array. Further acceleration is possible using user-defined sub-arrays, exceeding 600 fps from a 128 x128 sub-array, all whilst maintaining single photon sensitivity and quantitative stability throughout. The status of ‘Ultimate Sensitivity’ is also preserved in this model, with thermoelectric cooling down to -80°C and industry-lowest clock induced charge noise. Additional unique features of the iXon include USB connectivity and iXon Ultra models also provide direct raw data access for on the fly processing. EMCCD and conventional CCD readout modes provide heightened application flexibility, with a new ‘low and slow’ noise performance in CCD mode.
Explore our related assets below...
