Resources
 Part of the Oxford Instruments Group
Part of the Oxford Instruments Group
Expand
Collapse
 Part of the Oxford Instruments Group
Part of the Oxford Instruments Group
Fluorescence microscopy enables the study of cells with molecular specificity and subcellular resolution. Despite recent improvements in the brightness of fluorescent labels and in the sensitivity of image sensors, imaging is usually limited to stationary cells. Imaging fluorescence in moving cells is fundamentally challenging because the exposure time is constrained by motion-blur, which limits the available signal. Ethan Schonbrun and his colleagues at Harvard University have developed a method that enables fluorescence imaging of fluorescently labelled cells traveling at high linear velocities in fluids. Flowing cells are illuminated with a pseudo-random excitation pulse sequence, resulting in a motion-blur that can be computationally removed to produce near diffraction-limited images. With this method, sub-cellular and organelle structure can be studied under hydrodynamic forces for the first time. The decoded images are nearly diffraction limited even though the effective exposure time can be more than 50 times greater than a traditional motion-blur limited exposure. By using a coded excitation sequence, this system can image fast moving cells with a spatial resolution that is comparable to what is obtained when imaging fluorescence in stationary cells.
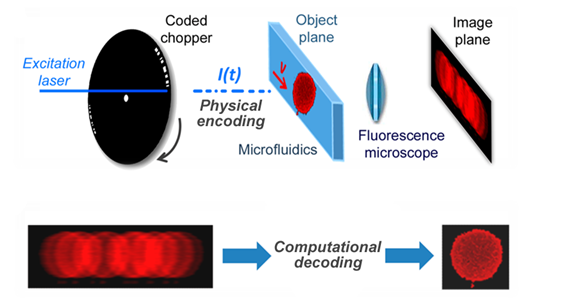
Figure 1. Coded excitation fluorescence microscope.
The image acquisition process is broken up into two steps, consisting of physical encoding and computational decoding of an engineered motion-blur. Instead of physically minimizing motion blur, a computational method to eliminate it was investigated. Based on the assumption that the imaging process is linear and shift-invariant, motion-blur can be removed by deconvolution. To implement a fluorescence imaging platform based on an engineered motion-blur, the system shown in Fig. 1 was developed by Schonbrun. The excitation laser has a wavelength of 488 nm and an optical power of 50 mW that is focused to a 160 µm full-width-at-half-maximum spot?size inside the microfluidic device. The camera is an Andor Neo scientific CMOS camera operated in global shutter mode. Physical encoding of the excitation light is performed by a custom -made chopper wheel, which modulates the fluorescence excitation with a temporal code that is a pseudo-random sequence of 18 pulses. Two coded sequences are produced for every rotation of the chopper wheel. When the chopper is spun at 100 Hz each pulse is 30 µs long, producing a total effective exposure time of 540 µs and the code has?a 960 µs total duration. Assuming a spatial resolution of 1 µm, the motion-blur limited velocity of a single pulse is 33 mm/s. Cells flow through a microfluidic channel that is 30 µm wide and 8 µm deep. While velocity may vary from cell to cell, due to laminar flow in the microfluidic channel, the velocity of an individual cell stays constant during illumination and the trajectory follows the fluid streamlines.
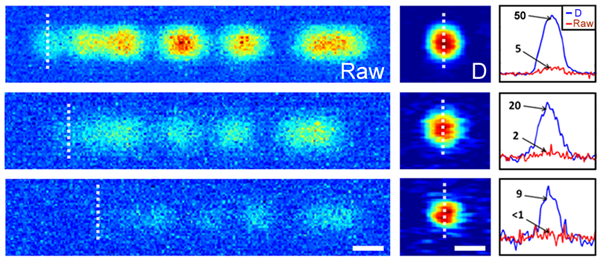
Fig 2. Motion Blur Decoding
To explore image reconstruction using a coded excitation, fluorescent beads traveling through a microfluidic channel were imaged. Figure 2 shows both raw and decoded images of 2 µm beads at four progressively decreasing excitation intensities. In all four cases, the bead is traveling at approximately 30 mm/s from left to right. The code modulates what would otherwise be a flat blur streak, approximately 30 µm in length. As can be seen from Fig. 2(e), even at an SNR below 1, the image can be reconstructed with reasonable fidelity. The code sequence, shown in Fig. 2(a), begins with an individual pulse. Because this pulse is isolated from the rest of the code by the two zeros that follow, the beginning of the motion-blurred image can be used to represent a single pulse exposure. This region of the motion-blurred image is denoted by a dashed line. We find that for all four excitation intensities, the reconstruction has an SNR that is approximately 10 times greater than that of the single pulse. This enhancement can be explained by a combination of the longer physical exposure and noise suppression from the deconvolution filter. While there is a trade-off between spatial resolution and background noise suppression in the implementation of the deconvolution filter, the longer effective exposure time enabled by the coded exposure increases signal strength without this trade-off. Schonbrun further extended this imaging approach to fluorescently labeled cellular organelles and found that it was possible to observe the relative location and morphology of these organelles in fast moving fluids. Motion-blur was effectively removed in this experiment, and intensity variations inside the nucleus were clearly resolved.
Fluorescence imaging of cells traveling in flow could enable image based cell sorting and high-throughput microscopic characterization of extremely large numbers of cells. In addition, non-adherent cells could be imaged in conditions similar to the blood stream where hydrodynamic forces are known to play an important role in cell morphology and function. As well as this, the method can be used for the study of the dynamics of blood and cancer cells in conditions that resemble the vasculature.
Fluorescence imaging of flowing cells using a temporally coded excitation; Optics express; February 2013; Vol 21, No.4. Ethan Schonbrun, Sai Siva Gorthi, Diane Schaak
Andor Solutions for Imaging cells in flow
Due to the fast movement of cells in this particular application the global shutter mode of the Neo and Zyla 5.5 is required. When rolling shutter mode was utilised, Schonbrun noticed the cell shape was distorted. The Neo 5.5 and the Zyla 5.5 have a 5.5-megapixel sensor that offers a large field of view and high resolution without compromising read noise or frame rate. The small pixel size of 6.5 micrometers ensures sufficient oversampling of the point spread function, even for objectives with low magnification (e.g. 10x NA 0.3; 20x NA 0.5). The read noise is very low, even when compared with the highest-performance slow readout CCDs. The Neo 5.5 and Zyla 5.5 can achieve a read noise down to 1 e- without signal amplification, while reading out 5.5 megapixels at 30 & 100 frames per second respectively for the Neo 5.5 and Zyla 5.5 thereby facilitating the acquisition of dynamic events such as the imaging of fast moving cells in flow.
