Resources
 Part of the Oxford Instruments Group
Part of the Oxford Instruments Group
Expand
Collapse
 Part of the Oxford Instruments Group
Part of the Oxford Instruments Group
Lampe et al have developed a novel variant of direct STORM (dSTORM) termed spectral demixing dSTORM (SD-dSTORM) that combines the photochemical advantages of the red emitting carbocyanine dyes with the principal of spectral demixing. Specifically, they use a novel combination of carbocyanine dyes for super resolution, Alexa Fluor 647 and Alexa Fluor 700, which both show the excellent buffer compatible blinking characteristics needed for single molecule localization. SD-dSTORM requires reduced laser power and fewer imaging frames for faithful super resolved reconstruction of linear and punctuate biological nanostructures compared to other super resolution techniques.
Amongst the currently available organic photoswitches, the carbocyanine dyes Cy5 and Alexa 647 are most efficient for single molecule localization. This is based on their photostability (up to 6,000 photons per ON cycle) and, most importantly, their ability to exhibit a prolonged OFF state in a reducing environment. Lampe et al wanted to use Alexa 647 and needed to find a fluorescent partner that was both buffer compatible and one that exhibited a prolonged OFF state. Alexa 700 was chosen as the candidate. It was found that Alexa 700 exhibited the prolonged OFF state when in 100 - 300 mM β-mercaptoethylamine (MEA) and oxygen scavenger, and when excited at 643 nm. To distinguish the overlapping emission spectra of Alexa 647 and 700 the Optosplit II, a dual channel emission splitter from Cairn Research, was employed. The emission wavelengths were split via a dichroic mirror (710 DCXR) and two emission filters, HC 687 / 40 and ET 794 / 160, into short and long wavelength emission, and detected side by side on Andor’s iXon 897 EMCCD. The dichroic and bandpass emission filters for each channel were matched to the single laser line (643 nm) and the emission spectra to optimize the cross-talk required for spectral demixing.
To evaluate the quality of the spectral separation of Alexa 647 and Alexa 700, microtubules were labelled in BS-C-1 cells with commercially available secondary antibodies separately for each color, and mounted in dSTORM buffer (MEA, O2 scavenger buffer). Before acquisition, the sample was illuminated with 3 - 5 kW/cm2 at 643 nm to drive the fluorophores into the OFF state until the microtubule structure dissolved and stochastic blinking of the dyes was observed. Typically, 5,000 - 20,000 frames were acquired with a continuously running EMCCD at the same excitation. After acquisition, the single molecule localizations and their individual intensity values over the whole dual-channel view were determined with the open source software rapidSTORM. Lampe et al used their own custom written algorithm to get the fully reconstructed dualcolor SD-dSTORM image.
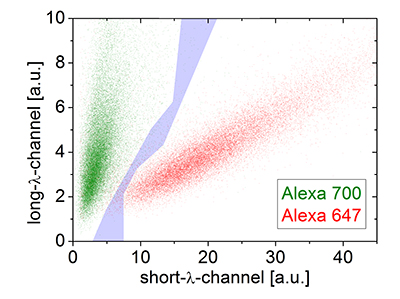
Figure 1. SD-dSTROM reconstruction of microtubules. Microtubules were stained separately with either Alexa 647 (red) or 700 (green) and imaged with SD-dSTORM. The intensity values for localization of Alexa 647 (red) and Alexa 700 (green) show different populations in a 2D intensity histogram. The intensity-based color assignment filter discards localizations in the cross-talk region (grey).
To determine the experimental optical resolution of the two color SD-dSTORM system, the scattering of single molecule localizations in SD-dSTORM was imaged (Figure 1). From this analysis the localization clusters appeared as fully separate spots, demonstrating the low probability of cross-talk between channels. The resolution for both channels, 22 nm for Alexa 647 and 30 nm for Alexa 700, were very similar to previously reported resolution values using Alexa 647 as a fluorophore for localization microscopy.
The applicability of SD-dSTORM in cell biology was demonstrated by imaging sub-cellular objects that are known to display distinctive shapes and spatial localization with different secondary antibodies labelled with Alexa 647 and Alexa 700. Focal adhesions, large protein complexes in the cell periphery, and clathrin coated pits, approximately 150 nm sized vesicular objects at the plasma membrane were chosen. Both do not co-localize with each other or with microtubules and should therefore appear as separate subcellular structures. Dual-color SD-dSTORM showed well separated super-resolved microtubules and focal adhesions (Figure 2A) and well separated microtubules and clathrin coated pits (Figure 2C). As a control, microtubules were labelled with both Alexa 647 and Alexa 700 to show the co-localization of the two channels (Figure 2B).
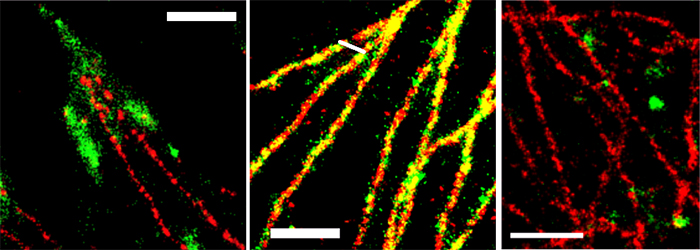
Figure 2. SD-dSTORM reconstructions of separate and co-localizing structures. SD-dSTORM of microtubules stained with Alexa 647 (red) and focal adhesion (A) or clathrin heavy chain (C) or microtubules (B, for colocalization test) stained with Alexa 700 (green). Scale bars = 1 μm.
Lampe et al have shown a significant advance towards user-friendly multi-color single molecule-based super resolution microscopy; it combines advantages of the red emitting carbocyanine dyes with the principle of spectral demixing to perform efficient, reliable and fast multi-color dSTORM. On the basis of spectral demixing, SDdSTORM offers the possibility of being extended to more than two colors using other related red dyes (i.e. Alexa 750). SD-dSTORM can be combined with any of the commercially available localization software (i.e. QuickPALM and rapidSTORM). SD-dSTORM promises to be an advantage for live cell imaging if combined with tag technology.
The setup for multi-colour direct STORM in the lab of Jan Schmoranzer and Andre Lampe showing the iXon3 897 and the OptoSplit II.
Research Paper: Multi-color direct STORM with red emitting carbocyanines, Biology of the Cell (2012), DOI: 10.1111/boc.201100011.
